Transcriptome analysis was performed to detect genetic modifications underlying citrus canker disease infected different citrus cultivars. Sequenced messenger RNA was analyzed using a suite of bioinformatic tools, based on a pipeline depicted on figure below. This pipeline initially analyze the RAW RNA-seq data for high quality reads recovery after a filtering step. Resulting reads are then mapped to citrus reference genome/transcriptome producing SAM alignments files. SAM alignments are then subjected to mutation detection for SNP and SSR repeats discovery, and to digital gene expression analysis (DGE). DGE is performed to find statistical significant (p-value<0.05) gene expression modification between citrus plants infected by Xac when compared to controls.
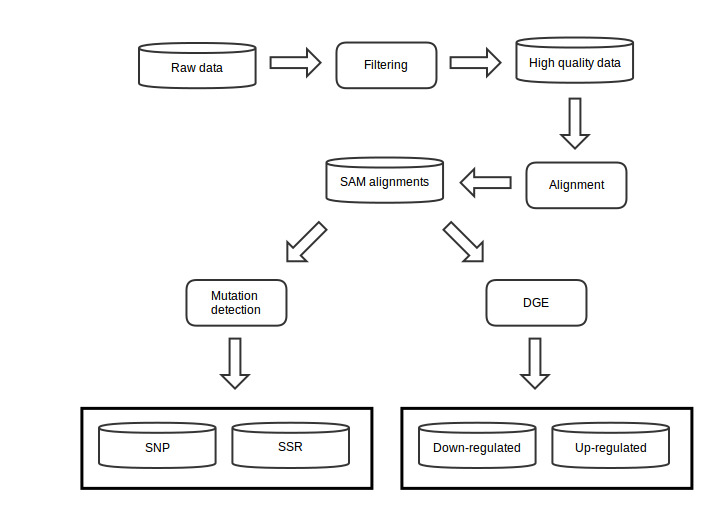
DGE genes can be induced (up-regulated) or repressed (down-regulated) genes during infection by X. citri. These DGE genes are then subjected to ontology analysis using Blast2GO software, to verify metabolic pathway alterations caused by X. citri.